Content
This document provide R script and data to reproduce the comparison between the spatial distributions of the observed and the estimated descriptors (states, fishing grounds and fishing intensity) for the variegated scallop dredging trip as illustrated in the supplementary figure S5 in Le Guyader et.al (2017):
- Le Guyader,D., C. Ray, F. Gourmelon, D. Brosset., (2017) - Defining high-resolution dredge fishing grounds with Automatic Identification System (AIS) data. Aquatic Living Resources. doi: 10.1051/alr/2017038
Data and replication
Rcode are available in this Github repository: https://github.com/dleguyader/LeGuyader-etal-2016-SM.
Load requires packages
# ipak function from Steven Worthington: install and load multiple R packages.
# https://gist.github.com/stevenworthington/3178163
# check to see if packages are installed. Install them if they are not, then load them into the R session.
ipak <- function(pkg){
new.pkg <- pkg[!(pkg %in% installed.packages()[, "Package"])]
if (length(new.pkg))
install.packages(new.pkg, dependencies = TRUE)
sapply(pkg, require, character.only = TRUE)
}
packages <- c("sp", "raster", "rasterVis", "rgdal", "rgeos", "spatstat", "maptools", "SDMTools", "adehabitatLT", "trip", "mclust", "ggplot2", "plyr", "dplyr", "caret", "viridis", "grid", "gridExtra")
ipak(packages)## sp raster rasterVis rgdal rgeos
## TRUE TRUE TRUE TRUE TRUE
## spatstat maptools SDMTools adehabitatLT trip
## TRUE TRUE TRUE TRUE TRUE
## mclust ggplot2 plyr dplyr caret
## TRUE TRUE TRUE TRUE TRUE
## viridis grid gridExtra
## TRUE TRUE TRUEPerform the analyses
1 - Import data and fix the parameters
######## Fix the parameters ################
dtm <- 600 # maximimum duration allowed between 2 positions(in seconds)
lhsv <- 47 # grid size: here we use the grid size caculated (in metres)
# for the 2011-2012 season(see paper for details)
maille <- 50 # smoothing factor (in metres): idem (see paper for details)
# Import data #######################
# GPS data for the variegated scallop fishing trip have been anonymized and
# coordinates have been modified with a vectorial translation for confidentiality matters
td <- getwd()
pet <- read.csv2(file = paste0(td, "/data/DataTripOneAnon.csv"))
coordinates(pet) <- ~coords.x + coords.y # To SPDF
proj4string(pet) <- CRS("+init=epsg:2154")
pet$time <- as.POSIXct(pet$time, tz = "UTC") # set time to POSIXct format
## Create the clipping polygon from the AIS data extent (e: extent)
e <- extent(pet) + 2000
CP <- as(e, "SpatialPolygons")
proj4string(CP) <- CRS("+init=epsg:2154")
## Get coastline data and clip to data extent
land <- readOGR(dsn = paste0(td, "/data"), layer = "land")## OGR data source with driver: ESRI Shapefile
## Source: "C:\Users\Damien\Documents\R\document\DLG_WEBSITE_ACADEMIC_EN-FR\Version_4\content\post\data", layer: "land"
## with 2617 features
## It has 2 fieldsland <- spTransform(land, CRS("+init=epsg:2154"))
land <- gIntersection(land, CP, byid = TRUE)
## Get sea shape data and clip to data extent
sea <- readOGR(dsn = paste0(td, "/data"), layer = "sea")## OGR data source with driver: ESRI Shapefile
## Source: "C:\Users\Damien\Documents\R\document\DLG_WEBSITE_ACADEMIC_EN-FR\Version_4\content\post\data", layer: "sea"
## with 1 features
## It has 1 fieldssea <- spTransform(sea, CRS("+init=epsg:2154"))
sea <- gIntersection(sea, CP, byid = TRUE) 2 - Estimate fishing activity
## Function for data cleaning: time filter
prep.fun <- function(x, dtmax, analyse) {
# Fonction to calculate ditance (dist) between consecutive points, time
# duration (dt), mean speed (vmoy) and deletion according to dtmax x :
# the SPDF dtmax: maximimum duration allowed between 2 positions(in
# seconds), analyse: name of the metier (character string)
df <- x
# conversion to ltraj class
d.ltraj <- as.ltraj(coordinates(df), df$time, id = paste(analyse))
ld.ltraj <- ld(d.ltraj) # ltraj to data.frame class
df$dist <- ld.ltraj$dist
df$dt <- ld.ltraj$dt
df <- subset(df, !is.na(df$dt)) # NA delete
df <- subset(df, df$dt < dtmax) ## dt>dtmax delete
df$vmoy <- df$dist/df$dt # Mean speed calculation
return(df)
}
pet.pt <- prep.fun(x=pet, dtmax=dtm, analyse="Varieg. scall")
densy <- densityMclust(pet.pt$vmoy)
densy$modelName # Optimal model## [1] "V"densy$G # Optimal number of cluster according to the BIC value## [1] 4CLUSTCOMBI <- clustCombi(densy)
# entPlot(CLUSTCOMBI$MclustOutput$z, CLUSTCOMBI$combiM, abc = 'normalized')
## No elbow in the NDE value, so we get the optimal number
## of cluster according to the BIC value
## Final classification
yclust <- Mclust(pet.pt$vmoy, modelNames = densy$modelName, G = densy$G)
## We get G = 4 as we see no elbow in the NDE value
## Get classification values
pet.pt$CLUST <- yclust$classification
## Set classification (1) for estimated fishing positions and (0) for
## estimated non fishing positions given the selected cluster(s)
fishSet.fun <- function(x, minClus, maxClus) {
# minClus: identifier for the minimum speed cluster (integer) maxClus:
# identifier for the maximum speed cluster (integer)
vmin.clust <- NULL
max.clust <- NULL
vmin.clust <- min(x$vmoy[x$CLUST == minClus])
vmax.clust <- max(x$vmoy[x$CLUST == maxClus])
x$estim <- 0 # default steaming
x[x$vmoy >= vmin.clust & x$vmoy <= vmax.clust, "estim"] <- 1 # Fishing
x$act_estim <- "NO" # Steaming (i.e No Fishing)
x[x$vmoy >= vmin.clust & x$vmoy <= vmax.clust, "act_estim"] <- "FISH" # Fishing
x$act_estim <- factor(x$act_estim)
return(x)
}
pet.pt <- fishSet.fun(pet.pt, minClus = 2, maxClus = 2)
rm(densy, CLUSTCOMBI, yclust)
confMat <- confusionMatrix(table(pet.pt$act_estim, pet.pt$act_obs)) # Confusion matrix3 - Identify Fishing grounds
## Data preparation for observed fishing positions
val.obs <- subset(data.frame(pet.pt), select = c(coords.x, coords.y, time, id, obs))
names(val.obs) <- c("x", "y", "date", "id", "statut")
## Data preparation for estimated fishing positions
val.est <- subset(data.frame(pet.pt), select = c(coords.x, coords.y, time, id, estim))
names(val.est) <- c("x", "y", "date", "id", "statut")
## Preparation for trip format coercion
fun.group <- function(x) {
x %>% arrange(id, date) %>%
mutate(gap = c(0, (diff(statut) != 0) * 1)) %>%
mutate(group = cumsum(gap) + 1)
}
val.obs <- ddply(val.obs, .(id, as.Date(date)), .fun = fun.group)
val.est <- ddply(val.est, .(id, as.Date(date)), .fun = fun.group)
val.obs$mmsi <- val.obs$id
val.obs$id <- paste(val.obs$mmsi, as.integer(as.Date(val.obs$date)), val.obs$group, sep = "_")
val.est$mmsi <- val.est$id
val.est$id <- paste(val.est$mmsi, as.integer(as.Date(val.est$date)), val.est$group, sep = "_")
## Get only fishing positions
val.obs <- val.obs %>%
filter(statut == 1) %>%
dplyr::select(x,y,date,id)
val.est <- val.est %>%
filter(statut == 1) %>%
dplyr::select(x,y,date,id)
## Trip format calculation
lenst.obs <- tapply(val.obs$id, val.obs$id, length)
## Delete non-consecutives fishing positions
val.obs <- val.obs[val.obs$id %in% names(lenst.obs)[lenst.obs > 2], ]
lenst.est <- tapply(val.est$id, val.est$id, length)
val.est <- val.est[val.est$id %in% names(lenst.est)[lenst.est > 2], ]
## Observed fishing positions to trip format
coordinates(val.obs) <- ~x + y
proj4string(val.obs) <- CRS("+init=epsg:2154")
trip.obs <- trip(val.obs, c("date", "id"))
## Estimated fishing positions to trip format
coordinates(val.est) <- ~x + y
proj4string(val.est) <- CRS("+init=epsg:2154")
trip.est <- trip(val.est, c("date", "id"))
pet.est <- trip.est
pet.obs <- trip.obs
rm(trip.est, trip.obs, val.obs, val.est)
## Function to compute KDE line density
fishGround.fun <- function(fishTrip, sea, grid, h) {
# fishTrip : fishing positions coerced to trip format sea: sea clip to extent of
# data grid, h: grid size and smoothing factor
# 1- Data preparation
owin.sea <- as(sea, "owin") # Window owin
bif <- raster(sea, resolution = maille) # Create template raster
projection(bif) <- CRS("+init=epsg:2154")
bif <- raster::mask(x = bif, mask = sea)
# 2 - compute KDE line density
t.obs <- fishTrip
t.obs$jour <- as.Date(t.obs$date)
vv.obs <- sort(unique(as.Date(t.obs$date)))
psp.obs.tx <- as.psp(t.obs) # coerce to psp format
psp.obs.tx$window <- owin.sea # affect owin
# KDE line density
dk.lx.obs <- density(psp.obs.tx, sigma = lhsv, dimyx = c(nrow(bif), ncol(bif)))
dk.obs <- raster(dk.lx.obs) # Raster conversion
projection(dk.obs) <- CRS("+init=epsg:2154")
return(dk.obs)
}
## Computation of KDE line density
pet.dk.obs <- fishGround.fun(fishTrip = pet.obs, sea = sea, grid = maille, h = lhsv)
pet.dk.est <- fishGround.fun(fishTrip = pet.est, sea = sea, grid = maille, h = lhsv)4 - Compute Fishing Intensity
## Function to compute the total fishing time spent per surface unit
fishInt.fun <- function(fishTrip, sea, grid) {
# fishTrip : fishing positions coerced to trip format sea: sea clip to
# extent of data grid size: see the paper
owin.sea <- as(sea, "owin") #Window owin
bif.trip <- raster(sea, resolution = grid)
bif.trip[] <- round(runif(nrow(bif.trip) * ncol(bif.trip)) * 100)
bif.trip <- raster::mask(bif.trip, sea)
bif.trip.grid <- as(bif.trip, "SpatialGridDataFrame")
bif.trip.gt <- getGridTopology(bif.trip.grid) # Create empty grid
tripgrid <- tripGrid(fishTrip, grid = bif.trip.gt, method = "pixellate") # compute time spent fishing
time.t <- raster(tripgrid)
projection(time.t) <- CRS("+init=epsg:2154")
time.est <- raster::mask(time.t, sea)
return(time.est)
}
## Calculation of the total fishing time spent per surface unit
pet.time.obs <- fishInt.fun (fishTrip = pet.obs, sea = sea, grid = maille)
pet.time.est <- fishInt.fun (fishTrip = pet.est, sea = sea, grid = maille)5 - Perform comparison with Warren’s similarity index
## Function for Warren's similarity index
evalMeth.fun <- function(x, y) {
# x and y are rasters obs and estim
asc.est <- asc.from.raster(x)
asc.obs <- asc.from.raster(y)
# ensure all data is positive
asc.est = abs(asc.est)
asc.obs = abs(asc.obs)
# calculate the I similarity statistic for Quantifying Niche Overlap
I = Istat(asc.est, asc.obs)
print(I)
}
## Get Values
pet.ground.eval <- evalMeth.fun (pet.dk.obs, pet.dk.est)## [1] 0.9609129pet.intens.eval <- evalMeth.fun (pet.time.obs, pet.time.est)## [1] 0.92329376 - Plot the figure
basemap <- list("sp.polygons", land, fill = "gray90", col = "gray60", lwd = 1)
north <- list("SpatialPolygonsRescale", layout.north.arrow(type = 1),
offset = c(146100, 6835650), scale = 600)
scale <- list("SpatialPolygonsRescale", layout.scale.bar(), scale = 1500,
offset = c(152600, 6831350), fill = c("transparent", "black"))
text1 <- list("sp.text", c(152600, 6831650), "0")
text2 <- list("sp.text", c(154000, 6831650), "1.5 km")
ext.x <- c(extent(pet.pt)[1], extent(pet.pt)[2])
ext.y <- c(extent(pet.pt)[3], extent(pet.pt)[4])
valKappa <- list("sp.text", c(152600, 6831650),
paste0("Kappa value = ", round(confMat$overall[2], 2)))
p1 <- spplot(pet.pt, "act_obs", col.regions= c("red", 'transparent'),edge.col='grey80',
alpha=0.5,cex = 0.5,lwd=0.8,legendEntries = c("Fishing", "No fishing"),
key.space= "bottom",
par.settings=list(fontsize=list(text=9)),
sp.layout=list(basemap, north,scale, text1,text2),
main = list(label="Observed states",
font = 1,just = "left",x = grid::unit(5, "mm")))
p2 <-spplot(pet.pt, "act_estim", col.regions= c("red", 'transparent'),edge.col='grey80',
alpha=0.5,cex = 0.5,lwd=0.8,legendEntries = c("Fishing", "No fishing"),
key.space= "bottom",
par.settings=list(fontsize=list(text=9)),
sp.layout=list(basemap, valKappa),
main = list(label="Estimated states",
font = 1,just = "left",x = grid::unit(5, "mm")))
p3 <- levelplot(pet.dk.obs, xlim=ext.x,ylim=ext.y,margin=FALSE, xlab=NULL, ylab=NULL,
scales=list(draw=FALSE),col.regions = viridis, colorkey = list(space="bottom"),
main=list(label="Observed fishing ground",
font = 1,just = "left",x = grid::unit(5, "mm")),
par.settings=list(fontsize=list(text=9))) +
latticeExtra::layer(sp.polygons(land, fill= "gray90", col="gray50"))
p4 <- levelplot(pet.dk.est,xlim=ext.x,ylim=ext.y, margin=FALSE, xlab=NULL, ylab=NULL,
scales=list(draw=FALSE),col.regions = viridis, colorkey = list(space="bottom"),
main=list(label="Estimated fishing ground",
font = 1,just = "left",x = grid::unit(5, "mm")),
par.settings=list(fontsize=list(text=9))) +
latticeExtra::layer(sp.polygons(land, fill= "gray90", col="gray50")) +
latticeExtra::layer(sp.text (c(152600,6831650),
paste0("Similarity index I = ", round(pet.ground.eval, 2))))
p5 <- levelplot(pet.time.obs,xlim=ext.x,ylim=ext.y, margin=FALSE, xlab=NULL, ylab=NULL,
scales=list(draw=FALSE),col.regions = viridis, colorkey = list(space="bottom"),
main=list(label="Observed fishing intensity",
font = 1,just = "left",x = grid::unit(5, "mm")),
par.settings=list(fontsize=list(text=9))) +
latticeExtra::layer(sp.polygons(land, fill= "gray90", col="gray50"))
p6 <- levelplot(pet.time.est,xlim=ext.x,ylim=ext.y, margin=FALSE, xlab=NULL, ylab=NULL,
scales=list(draw=FALSE),col.regions = viridis, colorkey = list(space="bottom"),
main= list(label="Estimated fishing intensity",
font = 1,just = "left",x = grid::unit(5, "mm")),
par.settings=list(fontsize=list(text=9))) +
latticeExtra::layer(sp.polygons(land, fill= "gray90", col="gray50")) +
latticeExtra::layer(sp.text (c(152600,6831650),
paste0("Similarity index I = ", round(pet.intens.eval, 2))))
grid.arrange(p1,p2,p3,p4,p5,p6, nrow = 3, ncol = 2,
top= textGrob("Variegated scallops dredging trip \n",gp=gpar(fontsize=12,font=2)))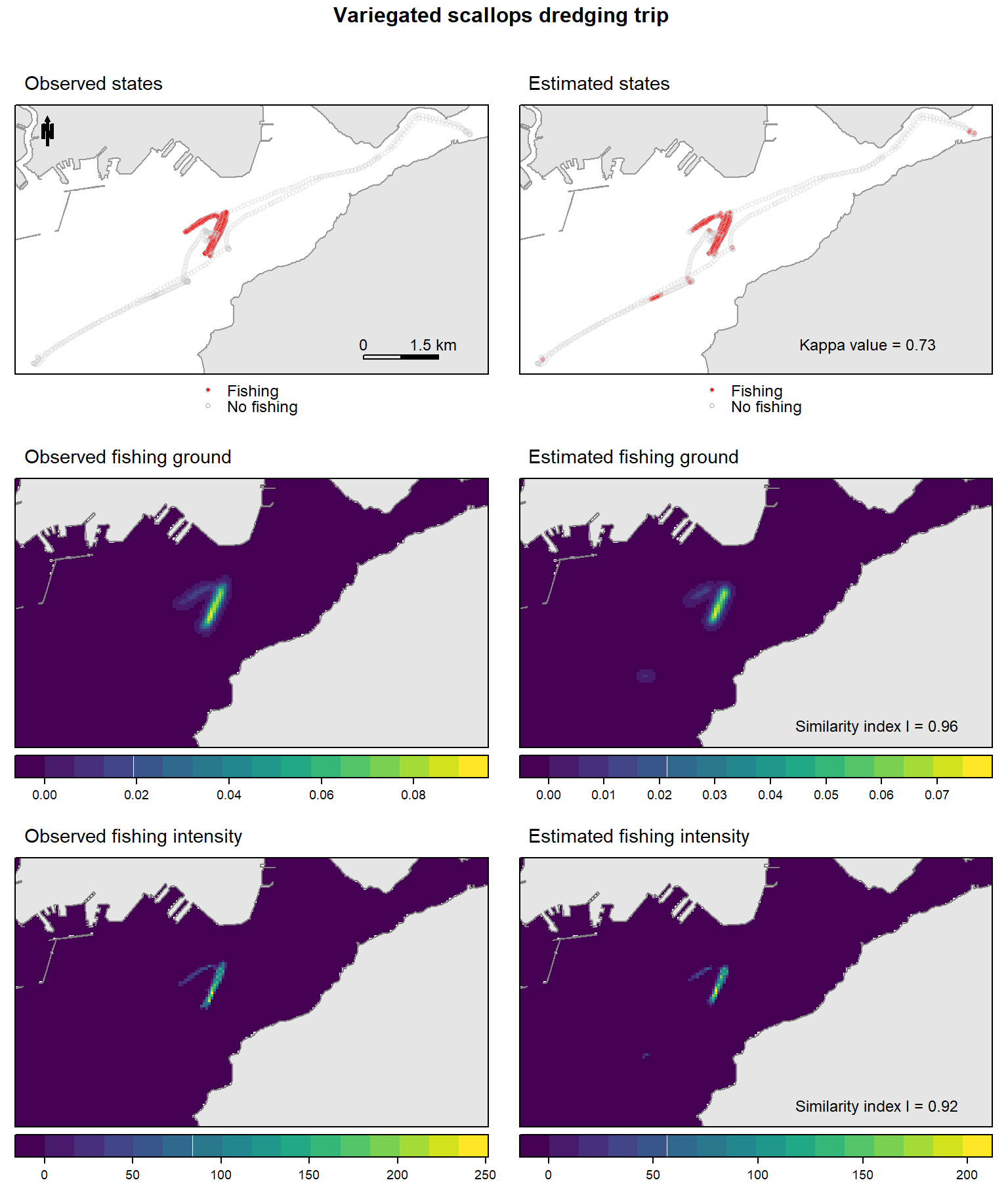
Figure 1: Spatial distributions of the observed (left) and the estimated (right) descriptors (states, fishing grounds and fishing intensity) for the variegated scallop dredging trip.